Seedlings S7 T100B (Sympathie X R.virginiana) X Lamarque



Seedlings S7 T152A (Sympathie X R.virginiana) X R.carolina



Seedlings S7 T152B (Sympathie X R.virginiana) X R.carolina
The two on the left hand side where crowded out by larger plants therefore smaller.



Great foliage Warren, any guess on color outcome from the crosses.
Thanks Dave
Probably pink or red, with the Lamarque cross I have no idea.
Cheers Warren
This seedling is a Hybrid Rugosa / Hybrid Virginiana
Seedling S5 T140 :- Fru Dagmar Hastrup (T) Type 2 X [Disco Lemonade X ( Sympathie X R.virginiana) X Elara)]
It has not flowered yet but the growth is vigorous and is now at 3 ft, foliage is very clean.


Seedling S102 T172 Dorotea X [(R. calocarpa x R. nutkana) x R. acicularis]. Dorotea is a (Sympathie X R.virginiana) X Elara.

A bit off topic, but I’m thinking I need to expand my species base, and was looking at either R. carolina or R. virginiana, and wondering about the relative merits of one over the other. If I’m not mistaken, Warren, you have worked with both, no?
I’ve been seeing more work being done with virginiana, and assume folks feel it has more to offer – would that be a fair assessment? May I inquire as to your or (not to hijack the thread) others relative experiences with these two species?
I treat them similarly and this is why.
Polyploid and hybrid evolution in roses east of the Rocky Mountainsl
Abstract
This study investigates the impact of hybridization and polyploidy in the evolution of eastern North American roses. We explore these processes in the Rosa carolina complex (section Cinnamomeae), which consists of five diploid and three tetraploid species. To clarify the status and origins of polyploids, a haplotype network (statistical parsimony) of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) nuclear gene was estimated for polyploids of the complex and for diploids of section Cinnamomeae in North America. A genealogical approach helped to decipher the evolutionary history of polyploids from noise created by hybridization, incomplete lineage sorting, and allelic segregation. At the diploid level, species west of the Rocky Mountains are distinct from eastern species. In the east, two groups of diploids were found: one consists of R. blanda and R. woodsii and the other of R. foliolosa, R. nitida, and R. palustris. Only eastern diploids are involved in the origins of the polyploids. Rosa arkansana is derived from the blanda–woodsii group, R. virginiana originated from the foliolosa–nitida–palustris group, and R. carolina is derived from a hybrid between the two diploid groups. The distinct origins of these polyploid taxa support the hypothesis that the three polyploids are separate species.
Wild species of roses are characterized by extensive morphological variation, which has resulted in a notoriously complex taxonomy. For instance, Linnaeus (Stearn, 1957⇓, p. 158) wrote in Species Plantarum, “The species of Rosa are with difficulty to be distinguished, with even greater difficulty to be defined; nature seems to me to have blended several or by way of sport to have formed several from one.” North American roses are no exception; Crépin (1896)⇓, Watson (1885)⇓, Rydberg (1920)⇓, and Erlanson MacFarlane (1966)⇓ described 13, 18, 129, and 22 Rosa species on this continent, respectively. Hybridization has long been considered to be one of the major causes of taxonomic confusion (Linnaeus, 1753⇓; Crépin, 1894⇓, 1896⇓), and artificial crosses have shown that in fact most diploids are interfertile (Erlanson, 1934⇓; Ratsek et al., 1939⇓, 1940⇓; Lewis and Basye, 1961⇓). Cytological studies during the early 20th century demonstrated that polyploidy is frequent in Rosa (Täckholm, 1922⇓; Hurst, 1925⇓) and that it could represent another source of variation. The present research explores issues related to hybridization and polyploidy, two important processes in plant evolution (Arnold, 1997⇓; Otto and Whitton, 2000⇓), that may explain the difficulty of recognizing species in wild roses.
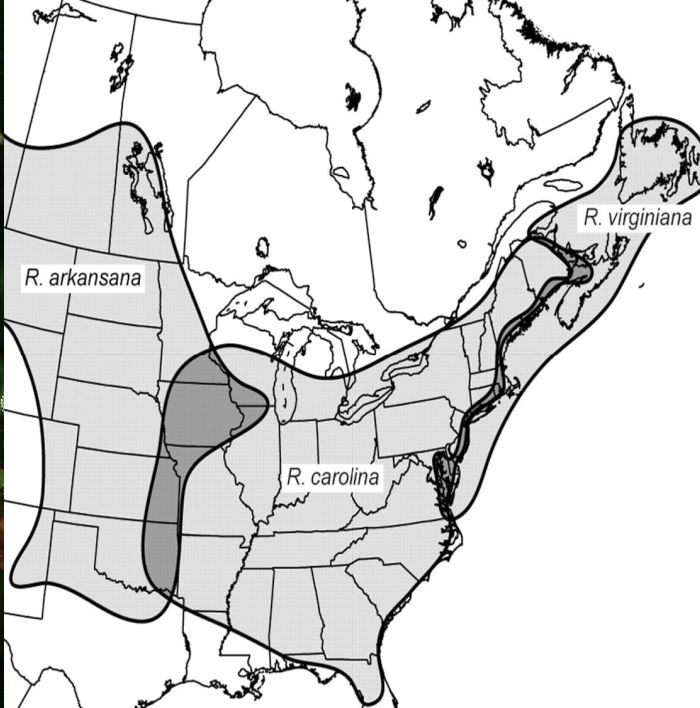
This study focuses on the North American Rosa carolina L. complex of section Cinnamomeae, a group that epitomizes the complexity of the genus. Indeed, Lewis (1957c⇓, p. 126) considered the group to be “… the most difficult taxonomic problem in our North American Rosa.” The complex consists of five diploid and three tetraploid species, almost entirely located east of the Rocky Mountains. The diploids R. blanda Ait., R. foliolosa Nutt., R. nitida Wild., R. palustris Marsh., and R. woodsii Lindl. (the sole species of the complex also found west of the Rocky Mountains) are relatively well circumscribed (Lewis, 1957c⇓; Erlanson MacFarlane, 1966⇓), but natural interspecific hybrids have been reported (Erlanson, 1929⇓, 1934⇓; Lewis, 1962⇓), and some have been given species status (Rydberg, 1920⇓; Erlanson, 1934⇓). In contrast, the tetraploid taxa R. arkansana Porter, R. carolina L., and R. virginiana Mill. are characterized by extensive continuous morphological variation that blurs their limits with each other and with their putative diploid ancestors in the R. carolina complex (Erlanson, 1934⇓; Lewis, 1957b⇓). Despite the important biosystematic investigations involving cytology and morphology in this complex (Erlanson, 1929⇓, 1934⇓; Lewis, 1957b⇓), the limits and origins of the polyploid taxa are still unclear. The broad polymorphism of polyploid species may be caused by hybridization given that it frequently has been reported in areas of contact between R. carolina and R. arkansana in the west (e.g., R. × rudiuscula Greene: Erlanson MacFarlane, 1966⇓; Lewis, 1957b⇓; A. Fishbein and W. H. Lewis, Washington University, unpublished manuscript) and between R. carolina and R. virginiana in the east (Fernald, 1922⇓; Lewis, 1957b⇓) (Fig. 1). Yet, it is also possible that these taxa represent a single polymorphic species rather than three distinct taxa. Therefore, reconstructing the origins of the polyploids is a logical first step toward a global understanding of the R. carolina complex because it could be relevant to solving the species status of the polyploids if these are shown to have evolved independently.
Fig. 1 Approximate distributions of the polyploid taxa Rosa arkansana, R. carolina, and R. virginiana. Areas where species overlap are in dark gray. The distributions are based upon Lewis (1957a)⇓ and personal collections
Several factors can impair our ability to determine from which species the polyploids evolved and whether they have evolved by autopolyploidy (from a single species) or by allopolyploidy (from more than one species: Grant, 1981⇓; Ramsey and Schemske, 1998⇓). For example, introgression of foreign alleles in an autopolyploid can hide its real origins by making it look like an allopolyploid. Other problems can result from irregularities in chromosome segregation. Allopolyploids are expected to have disomic segregation where chromosomes only pair with their homologues (bivalent formation) (Stebbins, 1950⇓, 1971⇓; Levin, 2002⇓), thus guaranteeing the preservation of homologous loci inherited from the parental species. However, these predictions are not always met, and allopolyploids may have occasional polysomic segregation via multivalent formation. This could lead to the fixation of alleles from a single parental species in the genome of the allopolyploid and hide its reticulate origin. The challenge when investigating polyploid evolution is thus to extract the true signal from the noise created by these confounding events in order to adequately reconstruct the evolutionary history of polyploids.
Investigation of polyploid origin must be done within a sound phylogenetic framework. To date, phylogenetic studies of Rosa have not included a good sampling of North American roses (e.g., Millan et al., 1996⇓; Matsumoto et al., 1998⇓; Wissemann and Ritz, 2005⇓), leaving their relationships obscure. Reconstruction of the diploid relationships could be further complicated by the recent origin of the complex, which is suggested by the low variation of ribosomal (Ritz et al., 2005⇓) and chloroplastic markers (Wissemann and Ritz, 2005⇓). Recent origin of species may result in incomplete lineage sorting of several molecular markers for the diploids (Pamilo and Nei, 1988⇓; Rosenberg, 2002⇓, 2003⇓), which in turn could hamper our ability to accurately identify the species that were involved in the origins of polyploids. These potential problems need to be addressed prior to investigating polyploid evolution.
A genealogical approach using a single-copy nuclear gene is used to address the relationship of diploids and to investigate the origins of the polyploids. A genealogical approach has major advantages over a genotyping method (e.g., microsatellites, amplified fragment length polymorphisms [ALFPs], isozymes) because it places the data in a historical perspective: it relates who is ancestral to whom rather than who is similar to whom. This is particularly important in order to discern some of the confounding events mentioned earlier from our principal goal—reconstructing polyploid evolution. The use of nuclear genes is particularly useful in this regard because non-haploid organisms (except for clonal and apomict taxa) receive one chromosome copy from each parent. Thus, nuclear genes can retain information about the reticulate history of organisms, which is impossible for maternally or paternally transmitted markers. Such an approach has been successful in reconstructing the polyploid origins of other taxa (Doyle et al., 2002⇓; Senchina et al., 2003⇓; Smedmark et al., 2003⇓; Helfgott and Mason-Gamer, 2004⇓; Joly and Bruneau, 2004⇓; Mason-Gamer, 2004⇓; Petersen and Seberg, 2004⇓; Evans et al., 2005⇓).
Very interesting article. Thank you, Warren. (Admittedly, at the third sentence I feared I was not going to be able to follow along!) Once the new species were created, I presume they evolved somewhat over their range
I found the Virginiana Hybrid H3 G40 (Red Single) very receptive to diploid pollen. This is seedling S7 T4 , (Sympathie X R.virginiana) X Lorraine Lee. Lorraine Lea is a Tea rose from the cross of (Jessie Clark X Captaine Millet).

Seedling S11 T161
[(Mimas X R.virginiana) X Freisia] X [ (Sympathie X R.virginiana) X Therese Bugnet]
Pretty healthy and armed to the teeth with thorns.


Seedling S35 T49

Seedling S5 T140 Hybrid Rugosa/ Hybrid Rosa virginiana
Seedling S35 T164 Hybrid Rosa virginiana X a seedling of [(R. calocarpa x R. nutkana) x R. acicularis]
Seedling S35 T137 Hybrid Rosa virginiana X (Sympathie X Eyes for You)



Seedling S32 T142 Hybrid Virginiana X Hybrid Rugosa
Seedling S21 T120 Hybrid Virginiana X Hybrid Rugosa


Hybrid China/ Hybrid Rosa virginiana

The foliage on many of these hybrids look absolutely spectacular! I love seeing what’s achievable with R. virginiana and its hybrids.
Ditto that! (The thorns on some of them, on the other hand… ) I haven’t been posting reactions, but I have been vicariously enjoying your seedlings Warren. Thanks for sharing!
Love the pictures Warren!
Thanks Andre, these hybrids I use seem to compatible with everything, producing good offspring which are very fertile.
Hi Phil; that seems to happen when you start adding a little Rugosa to the recipe.
Dank je Dane.